Menu
Frequently Asked Questions (FAQs)
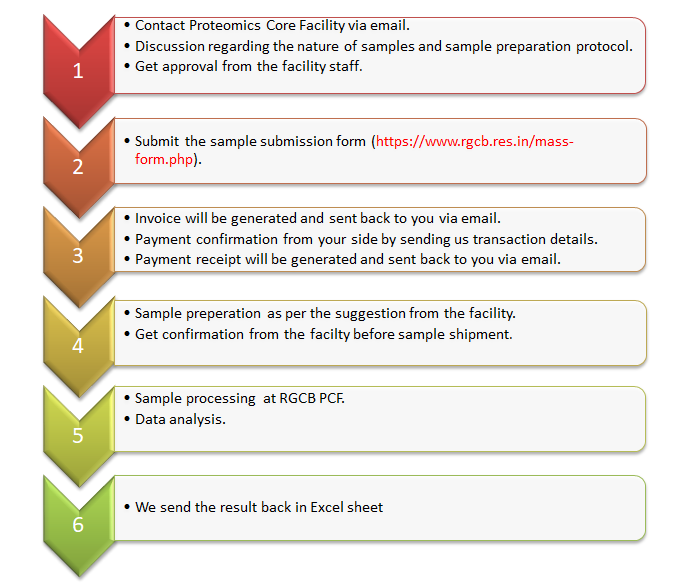
- To submit your samples, first, apply using the online sample submission form (proteomics link, metabolomics link).
- Once you submit the form, you will receive an email acknowledging the receipt of your application.
- After that, we will generate an invoice and will send it back to you for the costs of the mass spectrometry services. It usually takes 3-5 working days to generate the invoice.
- After receiving the invoice, you can make the payment according to the payment options (online, DD or direct cash) provided in the invoice.
- Once the payment is made, share the transaction details (reference number, date of payment) with us so that we will generate a payment receipt and will send it back to you.
- After making the payment, you can submit the samples at a convenient time after consulting with our facility (by email or telephone).
Please refer Guidelines
Please refer guidelines
-
In-solution protein samples: We prefer to have a concentration of 1 mg/mL and 100 µl is required (i.e., 100 µg in 100 µl). If the concentration of your sample is less than 1 mg/mL, let us know in advance. If you have samples to be compared (for relative protein quantification), all samples should be in uniform concentration and volume. In this case, we prefer the samples to be in aqueous solution of ammonium bicarbonate buffer (50 mM) with pH ~8, which is optimal for trypsin activity.
-
Gel bands: A visible band with Coomassie stain is sufficient. However, if the band is extremely faint, there is a chance that we may not get any identification. So, we suggest you load the maximum possible amount of protein solution into the polyacrylamide gel matrix so that we get a fairly stained gel band.
-
Molecular weight determination: We prefer to have a concentration not lower than 1 mg/mL and 20–50 µl is required. The buffer composition is not highly critical, as long as it does not contain detergents or more than 5% glycerol.
The common contaminants, even in small quantities, can mask important peaks in your MS data and have a huge impact on the final results. The usual contaminants in MS include keratin, polyethylene glycol, polypropylene glycol, phthalates, ions, polymeric detergents (tritons, tweens etc.) and siloxanes.
Controlling LC/MS contaminants
- Always use filtered de-ionized water.
- Perform sample processing in a laminar flow hood.
- Any glass container, flasks or tubes used to prepare buffers or samples must be thoroughly cleaned before use. Never use detergents or dishwasher to clean the beakers and glass plates. Always rinse glassware using organic solvents (70% ethanol or 70% methanol) and then filtered deionized water.
- Always wear particulate-free, powder-free, non-latex gloves and rinse them occasionally as they readily attract dust or hair particles. Avoid skin contact with the gloves on.
- Refrain from using deodorant and other cosmetic products during sample preparation. Siloxanes, present in these compounds, can cause MS contamination under certain conditions, such as nano flow.
- Do not store liquids in plastics (source of phthalates). Also, never use parafilm or other plastic films to cover solvent reservoirs.
- Visually inspect the vials, beakers, flasks, and tubes and make sure that they do not contain contaminants.
-
Detergent removal spin columns are available, for example, PierceTM Detergent Removal Spin Column which can be used to remove the detergents before sending in your samples to the facility.
-
Another way to remove detergents, salts, and other LC/MS incompatible things from samples is to do 'gel-assisted' proteolysis (Here, we trap the protein solution in a polyacrylamide gel matrix and wash out detergents, salts, and chaotropic agents, and perform in-gel digestion). In this case, run the protein sample approx. 1-2 cm into the gel. DO NOT allow the proteins to resolve in the gel (this is a sample clean-up strategy only). Then, PRECISELY cut out the entire sample up to the sample front (only the stained portion of the gel) - any excess gel will lead to background noise. Then, send us the diced gel band. We will perform in-gel digestion and subsequent LC/MS analysis.
-
Only volatile salts (e.g., ammonium bicarbonate, ammonium formate, ammonium acetate, triethylammonium bicarbonate (TEAB)) are compatible with mass spectrometers. Non-volatile salts and additives (e.g., sodium (Na+), potassium (K+), or phosphate) can precipitate at the high-organic end of the solvent gradient. This will block the columns and MS source, resulting in painstaking cleaning procedures. If your sample contain non-volatile salts or additives, perform dialysis, or use salt removal kits, for example c18 Zip Tip (Millipore, #ZTC18M096, 96 Pk), or a centrifugal filter with an appropriate molecular-weight-cut-off (MWCO) membrane. We suggest performing buffer exchange using ammonium bicarbonate buffer (50 mM), having pH around 8, using Amicon Ultra 0.5 mL centrifugal filters with 3,000 MWCO for this task (Millipore, #UFC500324, 24 Pk).
-
Another way to remove detergents, salts, and other LC/MS incompatible things from samples is to do 'gel-assisted' proteolysis (Here, we trap the protein solution in a polyacrylamide gel matrix and wash out detergents, salts, and chaotropic agents, and perform in-gel digestion). In this case, run the protein sample approx. 1-2 cm into the gel. DO NOT allow the proteins to resolve in the gel (this is a sample clean-up strategy only). Then, PRECISELY cut out the entire sample up to the sample front (only the stained portion of the gel) - any excess gel will lead to background noise. Then, send us the diced gel band. We will perform in-gel digestion and subsequent LC/MS analysis.
Please send duly packed samples to the following address:
Attention: Dr. Arun Surendran
Mass Spectrometry Core Facility
Second Floor (Extension: 4-536)
Rajiv Gandhi Centre for Biotechnology (RGCB)
Thycaud Post, Poojappura
Thiruvananthapuram - 695 014
Kerala, India
Ensure the samples arrive at RGCB on a weekday between 9:00 AM and 5:00 PM (Monday - Friday).
The results of the analysis would be communicated in the form of Excel sheets with data including protein accession number, protein name, peptide count, unique peptide count, molecular weight and abundance.
Where the position of a PTM is known/suspected, these can be scanned for directly using the analysis software. Enrichment strategies is needed for certain modifications (e.g., Phosphorylation). Please discuss PTM analysis with the facility staff.
Raw data is generally not sent with results. If you need the raw data, please contact the facility.
The raw data would be saved at the facility for a period of one year from the date of result communication.
Yes. The species details are always preferred as it would lead to precise data comparison using appropriate database.
In cases where the species (organism) of interest is not a model or widely studied organism, the user should either
provide a protein database (in .fasta format) or indicate a publicly available database for searching.
Please refer Service Charges
The "DBT-SAHAJ National Facility for Mass Spectrometry-based Proteomics, Metabolomics and Lipidomics at RGCB" must be acknowledged in publications and presentations.
A clear stained single gel band (usually) contains several proteins of (almost) the same molecular weight. Many of these proteins might be below the detection level of the staining method employed. The sensitivity and resolution of LC/MS is far superior compared to that of coomassie and silver staining.
There can be several reasons for this. The most common ones are given below.
- Your protein of interest might not be present in sufficient amounts to be detected among other proteins (of almost the same size) in the gel band.
- Your protein of interest might not have sufficient number of cleavage sites for trypsin, our protease. This usually happens for low molecular weight proteins.
- Your protein of interest is not present in the database. This usually happens when the species (organism) of interest is not a model or widely studied organism.
The biological variation of the samples is usually large, compared to the technical variation. Therefore, we suggest having multiple biological replicates for each sample (a minimum of two). We always do two injections per sample (two technical replicates).
Once we receive the samples, it generally takes 4-8 weeks to process them, conduct LC/MS analysis, and perform data analysis. However, this timeline may vary depending on the number of samples, our current backlog, and the availability of the LC-MS instruments. We will notify you if there are any delays in the processing of your samples.
